Backstory
On October 11, 2022, I received a rather interesting email from the Aggie Research Program, recruiting students for a project called FOODS (Fully Optimized Organically-Inspired Degradable Structures). In it, was a meme starring Forky, who would later go on to become the unofficial mascot of our project. Naturally, I applied, and a few weeks later, I was on my first ever research project.


FEA analyses of our partially-optimized fork
A Crash Course
The original team was composed of 4 undergrads - Archit, Roberto, Dian, and myself - none of whom had any experience with FEA (finite element analysis). So our advisors, Hannah and Jared (both PhD candidates under MAESTRO lab at the time), started us off with an FEA crash course.
We were introduced to a software called Abaqus and began learning how to go through the whole FEA pipeline - and eventually, how to automate it all using Python scripting. By the end of the Fall 2022 semester, I had put together a full-blown fork script - from modeling, all the way to analysis.
Research Begins
When we returned from winter break, it was time to begin the actual research. Essentially, our goal was to design a plastic fork with optimal biodegradability - by maximizing surface-area-to-volume ratio (SA:V) - while using FEA to ensure that the design wouldn't break.
Since I had progressed the furthest with my FEA skills, I stepped up to lead our effort. First, I defined a fork model with 44 parameters - the idea being that those parameters could be adjusted to find an optimal design. Using PLA as our material, I then created the parametrized fork in Abaqus, set up 2 critical load cases for analysis, generated meshes, and automated it with a Python script.


The meme that started it all
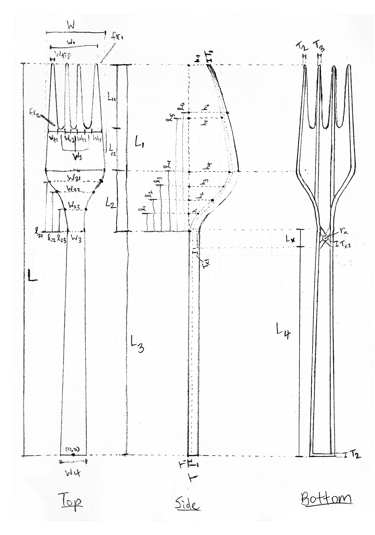
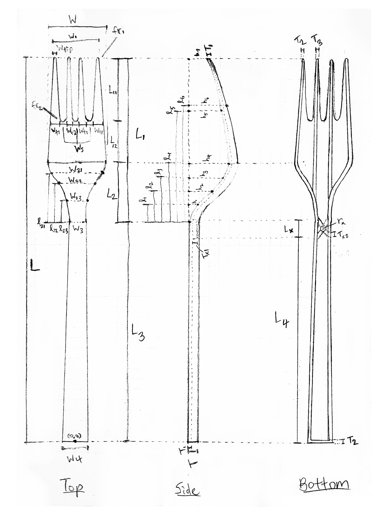
Now, optimizing all 44 parameters would have been impossible, so we had to first narrow it down to the ones with the greatest impact on SA:V and structural integrity. Through this DOE (Design of Experiments) process, I generated a slew of factor effects plots, ran a 2⁷ full factorial analysis, and ultimately, came up with 6 critical parameters.
Fork Optimization
With those critical parameters in hand, I could now start running optimization studies using scipy.optimize and try to find a fork that maximizes SA:V, while not failing under the specified load cases. In theory, it was a relatively simple optimization problem, but in practice, it was quite challenging.
The model and design space had to be constantly refined and improved upon. The optimization algorithm itself (Nelder-Mead) was gradient-based and required an initial guess, so there was the conundrum of choosing the right initial guess. Because if I didn't, then the algorithm would just find the nearest local maximum - not, in all likelihood, the global maximum that we were seeking.
The solution I came up with was to run a 3⁶ full factorial analysis, plus a differential evolution optimizer, in order to explore the design space thoroughly and figure out the best initial guess. Then, I could simply run that initial guess back through Nelder-Mead, and at last, we had our optimized fork - 3.4 times the SA:V of the COTS (commercial off-the-shelf) baseline fork that we had started with!
Parameterized fork sketch
Additional Work
Once we had our optimized design, I tried to 3D print it for experimental failure and degradation tests. Unfortunately, however, the only 3D printer that I had access to at the time was one that no one in the lab had gotten to work properly. I managed to get it working, through extensive trial and error, and even printed out several good replicas of our COTS baseline fork (as a control for the experiments). However, the optimized designs proved to be too difficult to print (due to their extremely low thickness).
I also tried to further optimize our design, using topology optimization in Tosca to figure out where additional material could be removed (i.e. adding holes to the fork). Through my analyses, I was able to identify the best locations for the holes, but I unfortunately never got around to running it through the full optimization pipeline again.
Priorities shifted as I tried to focus more on my aerospace-related projects, and though I stayed on for a while longer in an advisory role, I did eventually have to make the hard decision to leave the project.


COTS baseline vs partially-optimized fork
